Volume 12, Issue 3 (7-2024)
J. Pediatr. Rev 2024, 12(3): 273-282 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Hosseinzadeh F, Rahimzadeh G, Ahangarkani F, Salehpour S M, Rezai S, Valadan R, et al . The Emergence of Multi-drug-resistant and Extensively-drug-resistant Pseudomonas aeruginosa After the COVID‑19 Pandemic in North Iran. J. Pediatr. Rev 2024; 12 (3) :273-282
URL: http://jpr.mazums.ac.ir/article-1-647-en.html
URL: http://jpr.mazums.ac.ir/article-1-647-en.html
Fatemeh Hosseinzadeh1 

 , Golnar Rahimzadeh1
, Golnar Rahimzadeh1 

 , Fatemeh Ahangarkani2
, Fatemeh Ahangarkani2 

 , Seyedeh Mahsa Salehpour3
, Seyedeh Mahsa Salehpour3 

 , Shaghayegh Rezai4
, Shaghayegh Rezai4 

 , Reza Valadan5
, Reza Valadan5 

 , Laleh Vahedi6
, Laleh Vahedi6 

 , Somayeh Sheidaei6
, Somayeh Sheidaei6 

 , Faezeh Sadat Movahedi1
, Faezeh Sadat Movahedi1 

 , Raha Rezai1
, Raha Rezai1 

 , Mohammad Sadegh Rezai *7
, Mohammad Sadegh Rezai *7 




 , Golnar Rahimzadeh1
, Golnar Rahimzadeh1 

 , Fatemeh Ahangarkani2
, Fatemeh Ahangarkani2 

 , Seyedeh Mahsa Salehpour3
, Seyedeh Mahsa Salehpour3 

 , Shaghayegh Rezai4
, Shaghayegh Rezai4 

 , Reza Valadan5
, Reza Valadan5 

 , Laleh Vahedi6
, Laleh Vahedi6 

 , Somayeh Sheidaei6
, Somayeh Sheidaei6 

 , Faezeh Sadat Movahedi1
, Faezeh Sadat Movahedi1 

 , Raha Rezai1
, Raha Rezai1 

 , Mohammad Sadegh Rezai *7
, Mohammad Sadegh Rezai *7 


1- Pediatric Infectious Diseases Research Center, Communicable Diseases Institute, Mazandaran University of Medical Sciences, Sari, Iran.
2- Antimicrobial Resistance Research Center, Communicable Diseases Institute, Mazandaran University of Medical Sciences, Sari, Iran.
3- Students Research Committee, Mazandaran University of Medical Sciences, Sari, Iran.
4- Department of Medical Microbiology and Virology, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran.
5- Molecular and Cell Biology Research Center, Mazandaran University of Medical Sciences, Sari, Iran.
6- Department of Pathology, Faculty of Medicine, Mazandaran University of Medical Sciences, Sari, Iran.
7- Pediatric Infectious Diseases Research Center, Communicable Diseases Institute, Mazandaran University of Medical Sciences, Sari, Iran. ,drmsrezaii@yahoo.com
2- Antimicrobial Resistance Research Center, Communicable Diseases Institute, Mazandaran University of Medical Sciences, Sari, Iran.
3- Students Research Committee, Mazandaran University of Medical Sciences, Sari, Iran.
4- Department of Medical Microbiology and Virology, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran.
5- Molecular and Cell Biology Research Center, Mazandaran University of Medical Sciences, Sari, Iran.
6- Department of Pathology, Faculty of Medicine, Mazandaran University of Medical Sciences, Sari, Iran.
7- Pediatric Infectious Diseases Research Center, Communicable Diseases Institute, Mazandaran University of Medical Sciences, Sari, Iran. ,
Keywords: COVID-19, Drug resistance, Pseudomonas aeruginosa, Healthcare associated infections, Multiplex polymerase chain reaction (PCR)
Full-Text [PDF 549 kb]
(506 Downloads)
| Abstract (HTML) (1639 Views)
Full-Text: (323 Views)
Introduction
The COVID-19 pandemic has left an indelible mark on public health and healthcare management. One of the most pressing issues that has come to light is antimicrobial resistance (AMR). During the COVID-19 pandemic, 70% of patients with COVID-19 received antibiotics, either as outpatients or inpatients, which led to an increased risk of hospital-acquired infections and contributed to AMR following the pandemic. With the rise of infectious diseases and overuse of antibiotics, AMR has turned out to be a critical public health concern [1]. The COVID-19 pandemic has led to co-infections in many parts of the world, with a reported prevalence ranging from 0.35% to 53%. The widespread use of antibiotics to prevent or treat secondary infections in COVID-19 patients has further contributed to increasing antibiotic resistance. Patients with COVID-19 who are infected with Pseudomonas aeruginosa can transmit antibiotic resistance genes within healthcare facilities [1]. The irrational prescription of antibiotics and the surge in hospital admissions led to the rise of multidrug-resistant (MDR) and extensively drug-resistant (XDR) bacteria, such as P. aeruginosa. Infections caused by P. aeruginosa include pneumonia, urinary tract infections (UTI), as well as wound, ear and bloodstream infections. Moreover, P. aeruginosa is inherently resistant to many antimicrobial agents and is capable of developing secondary resistance to other available antibacterial classes. Strains of MDR and XDR P. aeruginosa are associated with a high death rate and pose a significant public health threat [2], due to the ineffectiveness of available treatment options against these strains, which have been identified as critical priority pathogens. Most risk factors for XDR P. aeruginosa are prevalent among patients with a prior hospital or ICU stay, those with prolonged use of broad-spectrum antibiotics and critically ill patients [3].
Beta-lactam antibiotics are highly effective and widely prescribed owing to their broad spectra and low toxicity. However, the emergence of mutated forms of beta-lactamases, such as extended-spectrum beta-lactamase (ESBLs)-producing isolates, AmpC beta-lactamases and carbapenemase genes, has become a major challenge to healthcare settings in treating infections. P. aeruginosa exhibits an outstanding ability to develop resistance to antimicrobial agents through chromosomal mutations and the acquisition of resistance genes, such as beta-lactamases, particularly carbapenemases, in addition to other resistance mechanisms [4]. As a result, there has been a rise in XDR and MDR strains.
Furthermore, this bacterium harbors numerous virulence factors that greatly contribute to its pathogenic nature, leading to the development of both acute and chronic infections. Researchers have identified several factors and characteristics that contribute to the pathogenicity of P. aeruginosa. This bacterium is capable of causing acute to chronic infections. These factors include cell-mediated factors, toxins and protease enzymes. P. aeruginosa can form biofilms, which is a significant virulence factor that helps it to survive in its host. Biofilm is a powerful defense mechanism that enables bacteria to resist antimicrobial drugs. It reduces the capacity for drug diffusion, creates low oxygen levels and allows dormant phenotypes to emerge. P. aeruginosa is a pathogenic bacterium that induces severe illness in humans.
One of its most potent virulence factors is the secretion systems it uses to inject toxic proteins into eukaryotic cells. The type III secretion system is particularly important, as it is responsible for injecting four toxic effector proteins—ExoU, ExoS, ExoT and ExoY—into the host cells’ cytoplasm. The presence of a functional type III secretion system is strongly correlated with poor prognosis and higher mortality rates. Approximately 70% of clinical strains of P. aeruginosa carry the gene that encodes ExoS, a factor that enhances the bacterium’s spread to the bloodstream and triggers host cell apoptosis. Additionally, the highly toxic virulence factor ExoA, which is secreted by the type 2 secretion system (T2SS), has been shown to cause severe damage to mammalian cells [5]. The ineffectiveness of current medications for P. aeruginosa infections is directly tied to the emergence of antibiotic resistance and the simultaneous presence of multiple virulence factors. This research was conducted to find the incidence of MDR and XDR P. aeruginosa isolated from patients with nosocomial infections (NIs) and investigate the presence of antibiotic-resistant genes in the COVID-19 era in Northern Iran.
Methods
This cross-sectional research across four teaching hospitals Bou Ali Sina, Fatemeh Zahra, Imam Khomeini and Shahid Zare in Sari, Iran, between May 2022 and June 2023. The research aimed to evaluate the incidence of MDR and XDR P. aeruginosa among patients with NIs, based on the national directory of NIs surveillance system. The study included various samples of hospitalized patients with NIs who contracted a new infection after three days of hospitalization. Additionally, it specifically focused on including MDR P. aeruginosa among the gram-negative isolates. Outpatients, as well as gram-positive and gram-negative isolates without multiple resistance, were excluded.
A laboratory technician collected various samples from 340 patients who were suffering from HAIs. The samples included urine, sputum, wound swabs, blood, cerebrospinal fluid (CSF), pleural fluid, eye swabs, endotracheal tube (ETT) samples, ear swabs and ascites. The samples were immediately sent to the central microbiology laboratory for analysis. As part of the routine procedure, all samples were cultured on MacConkey and blood agar (QUELAB, USA) plates. Blood specimens were inoculated in BD BACTEC plus Aerobic/F culture bottles (Becton Dickinson Company, Ireland). These bottles were then incubated in a BACTEC FX40 (BD, USA) culture system. Standard microbiological procedures were followed to identify P. aeruginosa [4, 5].
Demographic data were collected from the patients’ files (Table 1).
To test P. aeruginosa susceptibility, the standard broth dilution technique was used. Macro dilution trays were prepared with reagent-grade powders from manufacturers. The MIC of ampicillin-sulbactam, ceftazidime, cefepime, ciprofloxacin, colistin, co-amoxiclav, gentamicin, meropenem and piperacillin-tazobactam (Sigma, Germany) was determined to evaluate their efficacy. Strains resistant to at least one agent in three or more antimicrobial classes were described as MDR, while strains resistant to at least one agent in all but two or fewer antimicrobial classes were denoted as XDR. To combat the spread of antibiotic resistance, strains that produced ESBLs using the double-disk synergy test were screened. At least one of the following cephalosporins was used: Cefotaxime, cefepime, ceftazidime and ceftriaxone (Padtan Teb, Iran) to identify ESBL-producing strains. Additionally, the presence of ESBLs was detected using ceftazidime/clavulanic acid (30/10 μg), cefotaxime (30 μg), cefotaxime/clavulanic acid (30/10 μg), and cefepime/clavulanic acid (30/10 μg) antibiotic disks (Padtan Teb, Iran). The P. aeruginosa ATCC 27853 strain served as the positive control [6].
According to the company’s instructions, the DNA of bacteria was extracted using a commercial DNA extraction kit (Yekta Tajhiz, Iran). All isolates were screened by the multiplex PCR method for AmpC beta-lactamases, ESBL genes (blaCTX and blaSHV), MBL genes (blaSIM, blaSPM, and blaIMP), as well as ExoA and ExoS using specific primers as previously defined 4-5. The list of primers is presented in Table 1. The multiplex PCR reaction was prepared in a final volume of 20 μL, which included 10 μL of taq DNA polymerase 2x master mix RED, 1.5 mM of MgCL2 (AMPLIQON, Denmark), 0.5 μL of each primer (10 PM), 2 μL of DNA template (100 ng) and DNase-free distilled water. The amplification was performed using a Touchdown (TD) PCR protocol [7]. The following strains were used as positive controls: Klebsiella pneumoniae ATCC NO.51503 (blaCTX-M), K. pneumoniae ATCC NO. 700603 (blaSHV), Escherichia coli ATCC BAA NO.1143 (blaAmpC), Escherichia coli NCTC NO. 13476 (blaIMP), P. aeruginosa NCTC NO. 13921 (blaSPM) and P. aeruginosa NCTC NO. 14361 (blaSIM).
Statistical analysis
SPSS software, version 22 was used to analyze the data, employing descriptive statistics, chi-square tests, and Fisher’s exact tests.
Results
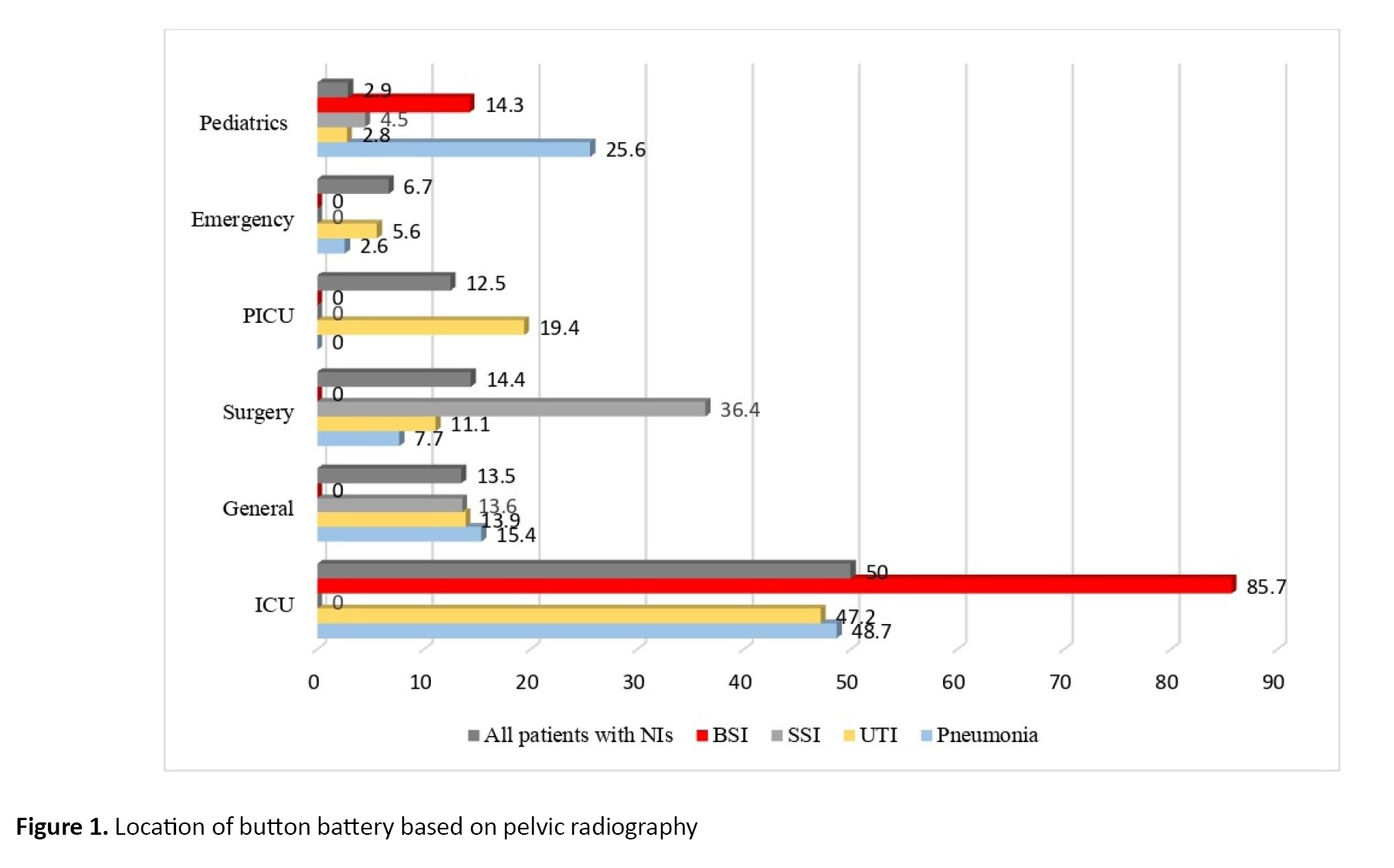
Out of 340 hospitalized patients with NIs caused by drug-resistant gram-negative bacterial pathogens, MDR and XDR P. aeruginosa were responsible for 104 cases (30.6%) of NIs. The median age of the patients was 58 years (IQR: 27–69.25 years), of whom 68 (65.4%) were male and 36 (34.6%) were female. The most common NIs caused by P. aeruginosa were pneumonia (39 cases; 37.5%), UTI (36 cases; 34.6%), surgical site infection (SSI) (22 cases; 21.2%) and bloodstream infection (BSI) (7 cases; 6.7%) (Table 1). The incidence of different types of NIs caused by MDR P. aeruginosa in various hospitalization wards was significantly different, as follows: ICU (52 cases; 50%), surgery (15 cases; 14.4%), general (14 cases; 13.5%), pediatric intensive care unit (PICU) (13 cases; 12.5%), emergency (7 cases; 6.7%) and pediatrics (3 cases; 2.9%) (P=0.002). The NIs caused by MDR and XDR P. aeruginosa in different wards are shown in Figure 1. Notably, 21.2% of strains isolated from patients with NIs (40.9% pneumonia, 40.9% UTIs, 13.6% SSIs and 4.5% BSIs) exhibited an XDR phenotype. Overall, 90.4% of isolates were ESBL-producing P. aeruginosa. Table 2 summarizes the in vitro antibiotic susceptibilities, including the MIC50, MIC90, geometric means (GM) MIC, and mode of MICs for ampicillin-sulbactam, ceftazidime, cefepime, ciprofloxacin, colistin, co-amoxiclav, gentamicin, meropenem and piperacillin-tazobactam against MDR and XDR P. aeruginosa.
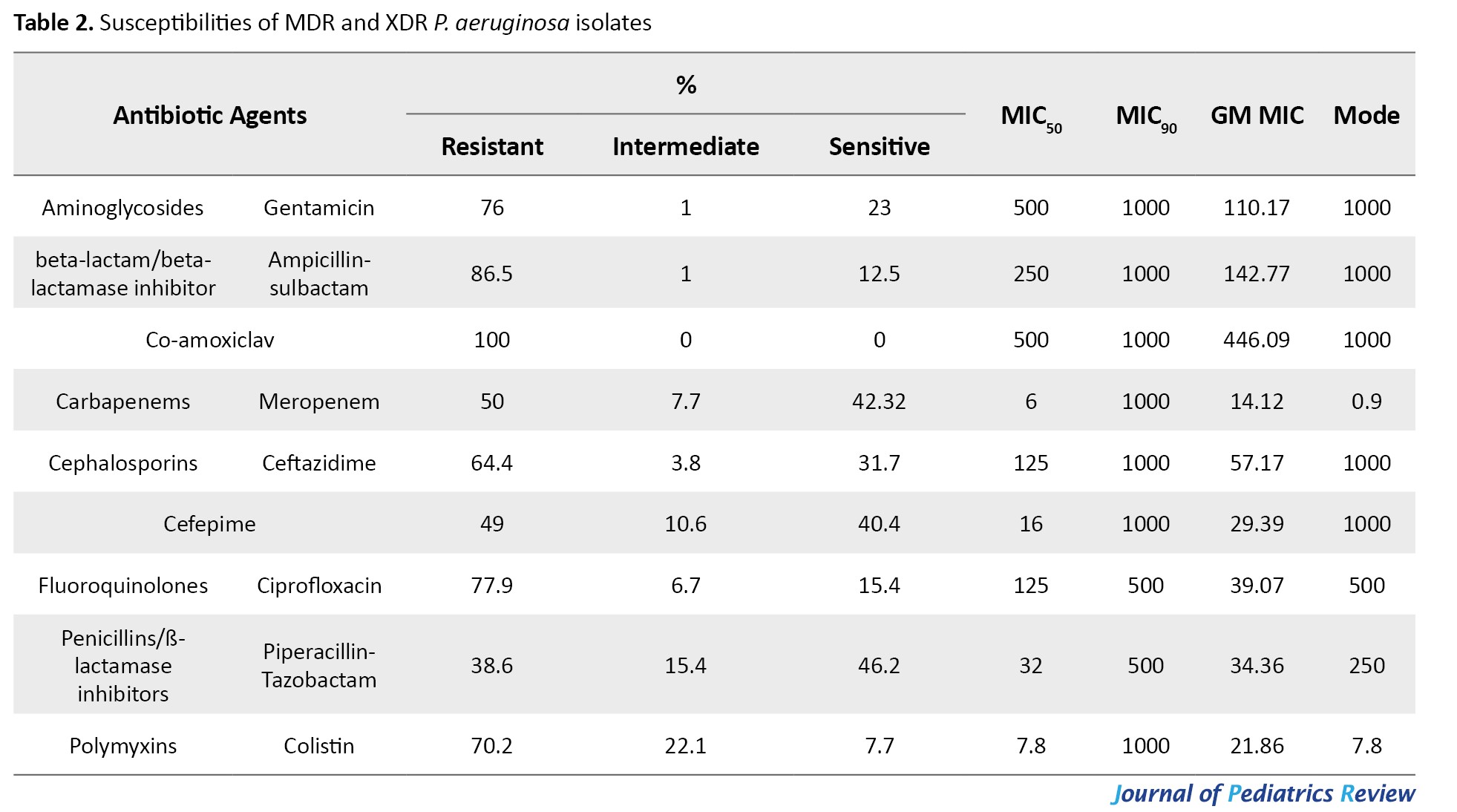
In terms of MIC50 values, co-amoxiclav, and gentamicin exhibited the lowest activity against P. aeruginosa. Regarding the geometric mean of minimum inhibitory concentration (GM MIC)values, meropenem exhibited the most potent activity, while co-amoxiclav displayed the lowest activity against most P. aeruginosa isolates. Also, the incidence of ESBL-producing P. aeruginosa among MDR and XDR isolates was 88.1% and 100%, respectively.
MIC50 and MIC90 represent the concentrations required to inhibit 50% and 90% of the isolates, respectively, while GM refers to the geometric mean.
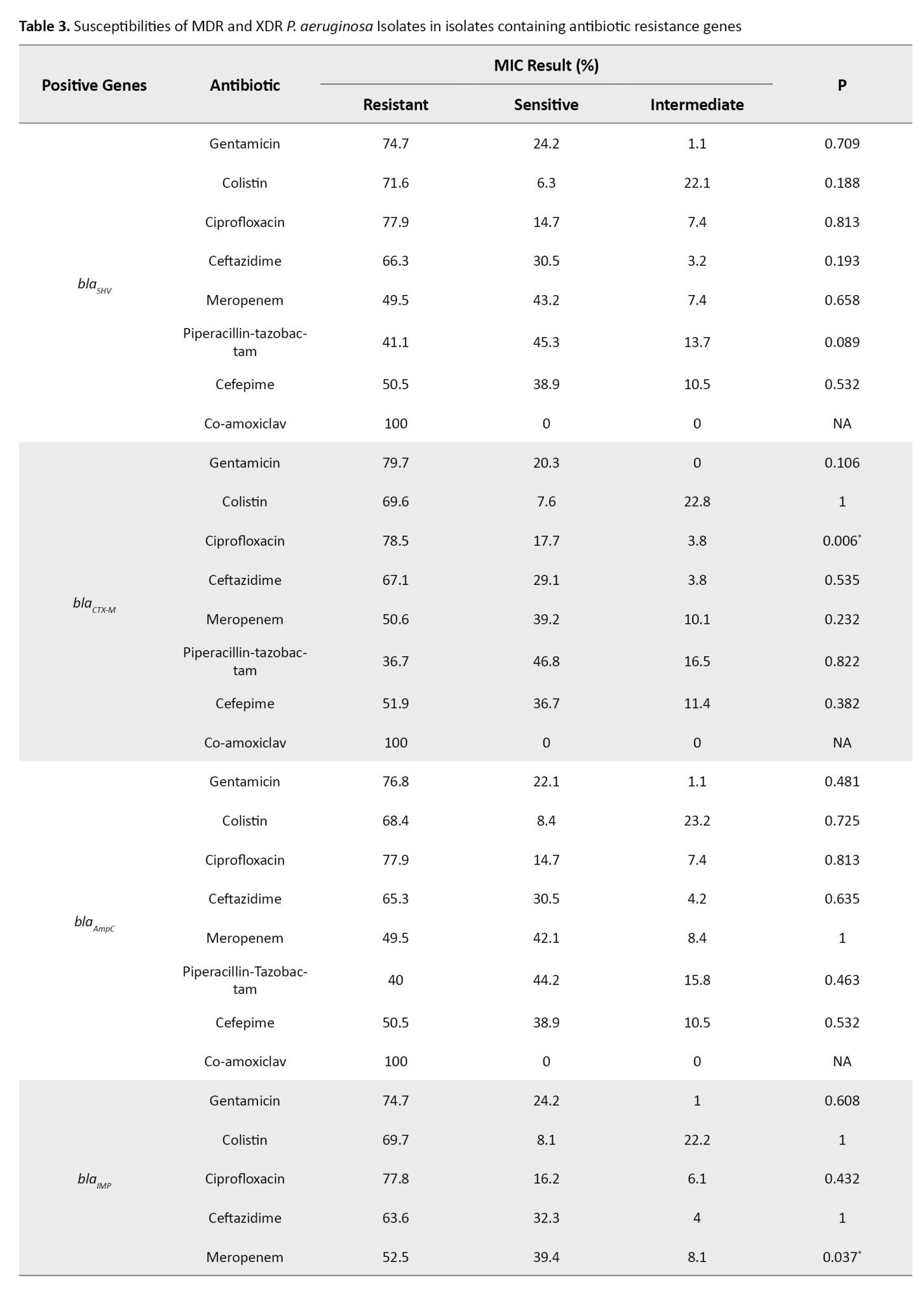
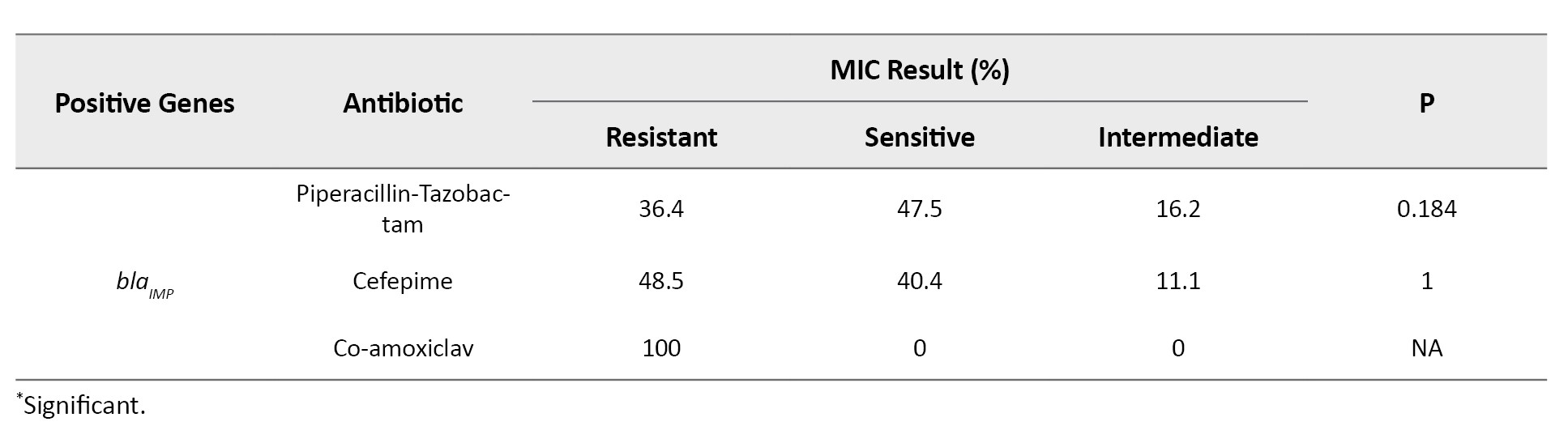
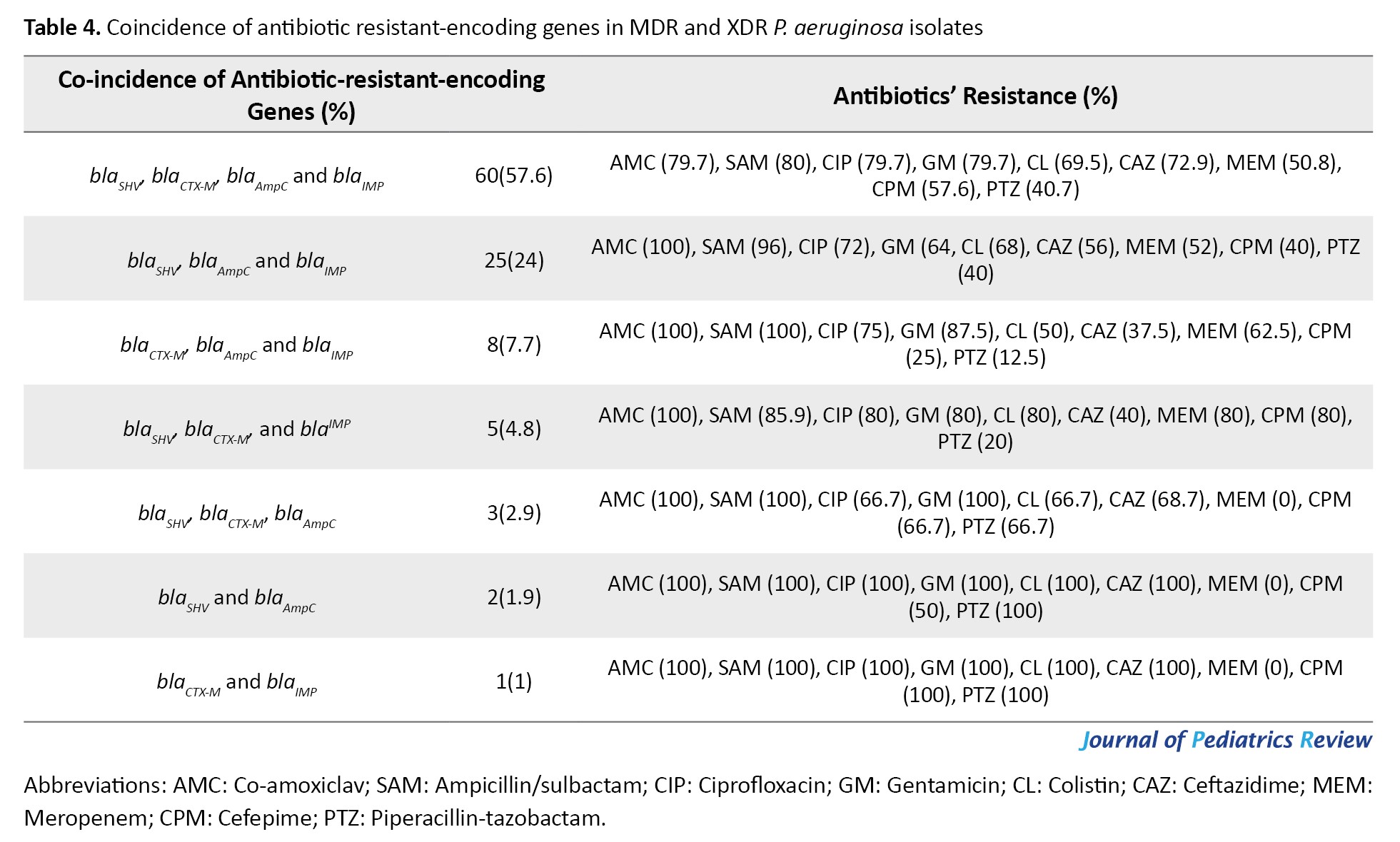
All isolates encoded ExoS gene, while the gene encoding ExoA was not observed among the isolates. The frequencies of resistance genes were as follows: blaSHV (91. 3%), blaCTX-M (76%), blaAmpC(91.3%) and blaIMP (95.2%), respectively. The blaSPM and blaSIM were not detected among the isolates. Based on antibiotic-resistant genes, seven genotypes were observed among the isolates. All isolates contained at least two antibiotic-resistant encoding genes. The most common genotypes included the co-presence of blaSHV, blaCTX-M, blaAmpC and blaIMP in 60 isolates (57.7%), blaSHV, blaAmpCand blaIMP in 25 isolates (24%), blaCTX-M, blaAmpC and blaIMP in eight isolates (7.7%), blaSHV, blaCTX-M and blaIMP in five isolates (4.8%), blaSHV, blaCTX-M, blaAmpCin three isolates (2.9%), blaSHV and blaAmpC in two isolates (1.9%), blaCTX-M and blaIMP in one isolate (1%), respectively. The blaCTX-M gene was significantly associated with resistance to ciprofloxacin (P<0.006) and the blaIMP gene was significantly associated with resistance to meropenem (P<0.03) (Tables 3 and 4).
 #T4
#T4
Discussion
P. aeruginosa is a major challenge in hospitals, causing high mortality rates, prolonged stays and increased costs [1, 2, 3]. The current study was carried out in the northern region of Iran and found that P. aeruginosa was accountable for 30.6% of NIs caused by MDR pathogens, with pneumonia accounting for the majority of these infections at 37.5%. Interestingly, previous surveillance studies conducted in teaching hospitals in the same area prior to the COVID-19 pandemic showed a significantly lower incidence of nosocomial pneumonia caused by ESBL-producing P. aeruginosa, at 14.63% [8]. Before the COVID-19 pandemic, UTIs (26.8%), VAP (20.3%), SSIs (19.7%) and BSIs (13.5%) were the most common NIs in different regions of Iran, according to a 2020 study [9]. The high occurrence of pneumonia in the current research is attributed to the high rate of ICU and PICU patients requiring invasive and non-invasive ventilation.
According to a 2014 survey, Pseudomonas spp. was the most commonly found bacteria in wound infections (50.81%), respiratory infections (21.31%), UTIs (19.67%) and blood infections (8.19%) [10]. However, a recent survey conducted in the post-COVID-19 era revealed that UTIs (33.33%), respiratory infections (32.46%), wound infections (19.30%) and blood infections (6.14%) were the most prevalent types of infections. In our study, MDR and XDR P. aeruginosa were responsible for 34.6% of UTIs, which is alarming because these infections are linked to severe forms of UTIs. They are often associated with urinary catheter-related infections, prostatitis and urolithiasis, which are very challenging to treat. Although P. aeruginosa is the third most common pathogen linked to nosocomial UTIs, the high incidence of UTIs caused by this bacterium is of particular concern [11].
SSIs account for 15% of all NIs in surgical patients, and SSIs caused by MDR bacteria are associated with significant postoperative complications and preoperative surgery [12]. The most common causes of SSI are Staphylococcus spp., Enterococcus spp., Streptococcus spp., and Pseudomonas spp. [13]. In this research, SSIs were caused by MDR and XDR P. aeruginosa in 22.2% of cases, consistent with another research [14]. Understanding the development and risk factors of SSI can help reduce treatment costs and allocate resources efficiently. In our study, the rate of BSI caused by MDR and XDR P. aeruginosa was 6.7%. There was also a link between AMR and poor clinical outcomes in patients with BSIs. Specifically, patients infected with MDR P. aeruginosa have twice the odds of mortality in comparison to those with non-MDR P. aeruginosa infections [15].
In a previous study involving patients with VAP and sepsis, it was discovered that approximately half of the isolated P. aeruginosa bacteria were resistant to all aminoglycoside antibiotics and 45.85% were resistant to ciprofloxacin. Moreover, the study found that P. aeruginosa exhibited resistance rates of 62.5% to colistin and 29.2% to imipenem [3]. After the COVID-19 outbreak, our recent surveillance investigations revealed an enhancement in resistance to aminoglycoside antibiotics among isolated P. aeruginosa bacteria, reaching 76%. Additionally, resistance to ciprofloxacin has risen to 77.9%. The resistance rates to colistin and meropenem among P. aeruginosa were found to be 70.2% and 50%, respectively.
Our results (2017) revealed a high incidence of catheter-associated UTIs in hospitals [16]. P. aeruginosa was recognized as a major cause of UTIs and demonstrated resistance to aminoglycosides (56%), fluoroquinolones (63%) and third-generation cephalosporins (38%). Following the COVID-19 outbreak, a further increase was observed in resistance to aminoglycosides (68.42%), fluoroquinolones (71.05%) and third-generation cephalosporins (68.42%) based on UTI samples. Tiri et al. found a significant increase in carbapenem-resistant Enterobacterales colonization from 6.7% (2019) to 50% (2020) [17].
During the pandemic period, AlDiba et al. discovered a substantial increase in the prevalence of carbapenem-resistant Enterobacterales, which rose to 22.4% from 5.4% in the pre-pandemic period [18]. The CDC’s latest report shows a 35% increase in MDR P. aeruginosa infections from 2019 to 2020, possibly due to the high administration of antibiotics to treat secondary bacterial infections associated with SARS-CoV-2 [19].
The rational usage of antibiotics during the COVID-19 pandemic was not followed. The antibiotics prescribed for COVID-19 patients were higher than the incidence of bacterial co-infections in these patients (bacterial co-infection [3.5%] and secondary bacterial infection [14.3%]) [20]. The high use of antibiotics in patients with COVID-19 during the pandemic raises concerns about the emergence of antimicrobial-resistant pathogens, such as resistant P. aeruginosa species, which have developed resistance to several antibiotic categories, such as cephalosporins, carbapenems, or polymyxins [1, 20]. In this study, a high resistance phenotype (38.5-100%) to different classes of antibiotics was observed, which is significantly higher than our previous finding (26-86.66%) before COVID-19 [3]. In the current study, piperacillin-tazobactam and imipenem were the most effective antibiotics against MDR and XDR P. aeruginosa. Our findings were in agreement with those of Ahmadi et al. who also reported that MDR P. aeruginosa was most susceptible to meropenem and piperacillin [21]. The antimicrobial sensitivity pattern in our examined strains showed higher resistance than P. aeruginosa isolates in other studies. This difference may be related to the studied population, the type of infections (as all strains were isolated from patients with NIs, not community-acquired infections) and the study period of three years after the COVID-19 pandemic.
The frequency of different ESBL genotypes varies in different regions. The fact that 57.6% of isolates in the current investigation contain all antibiotic-resistant genes (AmpC beta-lactamases, ESBL genes and MBL genes) is worrying. Several studies have reported the simultaneous presence of different β-lactamase genes in the same strains [22, 23]. The acquisition of drug-modifying enzymes in P. aeruginosa, such as ESBL and carbapenemases, can be accessed through horizontal gene transfer [4, 5]. Consistent with the current study, the blaSHV gene (86.66%) was the most detected ESBL gene in our previous survey [3]. Similar studies on ESBL-encoding genes revealed diverse findings [24]. The blaTEM, blaCTX and blaSHV variants have been the most common ESBLs during the past decade [25-27]. In this study, the most antibiotic-resistant-encoding gene was blaIMP, detected in 95.2% of isolates. However, 39.4% of isolates contained the blaIMP gene and showed a susceptible phenotype to meropenem. The incidence of blaIMP gene and resistance to meropenem was statistically significant (P=0.037). Most ESBL-encoding genes are plasmid-mediated enzymes, which are transmitted easily among bacteria, leading to inappropriate or failed antimicrobial therapy [28, 29]. Constant monitoring of bacteria carrying antibiotic resistance encoding genes (ESBL and MBL-producing strains) is pivotal to designating appropriate antimicrobial therapy.
Certain limitations in our study need to be taken into account. Firstly, the rate of hospital-acquired infections caused by MDR and XDR P. aeruginosa, as well as the presence of resistance genes, might have been underestimated due to partial treatment before the bacterial cultures were obtained in some cases. Secondly, we did not sequence the following genes associated with resistance phenotypes: blaSHV, blaCTX-M, blaAmpC and blaIMP, which limited our ability to match in vitro and in vivo resistance results. Despite these limitations, our findings provide crucial insights.
Conclusion
Our findings showed that UTIs, sputum and blood samples have the highest prevalence of blaSHV, blaCTX-M, blaAmpC and blaIMP genes in MDR P. aeruginosa isolates. The presence of these genes underscores the urgent need to expand empiric antibiotic therapy for critically ill patients. In addressing hospital-acquired infections caused by MDR bacteria, like carbapenemase and ESBL producers, it is imperative to implement suitable empiric or alternative treatments based on epidemiological data or relevant antibiotic exposure history. This study recommends the experimental use of meropenem to treat NIs caused by MDR P. aeroginosa in hospitals located in northern Iran.
Ethical Considerations
Compliance with ethical guidelines
This study was approved by the Ethics Committee of Mazandaran University of Medical Sciences (Code: IR. MAZUMS.REC.1401.13944).
Funding
This research did not receive any grant from funding agencies in the public, commercial, or non-profit sectors.
Authors contributions
All authors equally contributed to preparing this article.
Conflicts of interest
The authors declared no conflicts of interest.
Acknowledgements
The authors gratefully acknowledge Fatemeh Bagherzadeh, Ebrahim Nemati and Shahram Divsalar for their supportive assistance during the laboratory tests.
References
The COVID-19 pandemic has left an indelible mark on public health and healthcare management. One of the most pressing issues that has come to light is antimicrobial resistance (AMR). During the COVID-19 pandemic, 70% of patients with COVID-19 received antibiotics, either as outpatients or inpatients, which led to an increased risk of hospital-acquired infections and contributed to AMR following the pandemic. With the rise of infectious diseases and overuse of antibiotics, AMR has turned out to be a critical public health concern [1]. The COVID-19 pandemic has led to co-infections in many parts of the world, with a reported prevalence ranging from 0.35% to 53%. The widespread use of antibiotics to prevent or treat secondary infections in COVID-19 patients has further contributed to increasing antibiotic resistance. Patients with COVID-19 who are infected with Pseudomonas aeruginosa can transmit antibiotic resistance genes within healthcare facilities [1]. The irrational prescription of antibiotics and the surge in hospital admissions led to the rise of multidrug-resistant (MDR) and extensively drug-resistant (XDR) bacteria, such as P. aeruginosa. Infections caused by P. aeruginosa include pneumonia, urinary tract infections (UTI), as well as wound, ear and bloodstream infections. Moreover, P. aeruginosa is inherently resistant to many antimicrobial agents and is capable of developing secondary resistance to other available antibacterial classes. Strains of MDR and XDR P. aeruginosa are associated with a high death rate and pose a significant public health threat [2], due to the ineffectiveness of available treatment options against these strains, which have been identified as critical priority pathogens. Most risk factors for XDR P. aeruginosa are prevalent among patients with a prior hospital or ICU stay, those with prolonged use of broad-spectrum antibiotics and critically ill patients [3].
Beta-lactam antibiotics are highly effective and widely prescribed owing to their broad spectra and low toxicity. However, the emergence of mutated forms of beta-lactamases, such as extended-spectrum beta-lactamase (ESBLs)-producing isolates, AmpC beta-lactamases and carbapenemase genes, has become a major challenge to healthcare settings in treating infections. P. aeruginosa exhibits an outstanding ability to develop resistance to antimicrobial agents through chromosomal mutations and the acquisition of resistance genes, such as beta-lactamases, particularly carbapenemases, in addition to other resistance mechanisms [4]. As a result, there has been a rise in XDR and MDR strains.
Furthermore, this bacterium harbors numerous virulence factors that greatly contribute to its pathogenic nature, leading to the development of both acute and chronic infections. Researchers have identified several factors and characteristics that contribute to the pathogenicity of P. aeruginosa. This bacterium is capable of causing acute to chronic infections. These factors include cell-mediated factors, toxins and protease enzymes. P. aeruginosa can form biofilms, which is a significant virulence factor that helps it to survive in its host. Biofilm is a powerful defense mechanism that enables bacteria to resist antimicrobial drugs. It reduces the capacity for drug diffusion, creates low oxygen levels and allows dormant phenotypes to emerge. P. aeruginosa is a pathogenic bacterium that induces severe illness in humans.
One of its most potent virulence factors is the secretion systems it uses to inject toxic proteins into eukaryotic cells. The type III secretion system is particularly important, as it is responsible for injecting four toxic effector proteins—ExoU, ExoS, ExoT and ExoY—into the host cells’ cytoplasm. The presence of a functional type III secretion system is strongly correlated with poor prognosis and higher mortality rates. Approximately 70% of clinical strains of P. aeruginosa carry the gene that encodes ExoS, a factor that enhances the bacterium’s spread to the bloodstream and triggers host cell apoptosis. Additionally, the highly toxic virulence factor ExoA, which is secreted by the type 2 secretion system (T2SS), has been shown to cause severe damage to mammalian cells [5]. The ineffectiveness of current medications for P. aeruginosa infections is directly tied to the emergence of antibiotic resistance and the simultaneous presence of multiple virulence factors. This research was conducted to find the incidence of MDR and XDR P. aeruginosa isolated from patients with nosocomial infections (NIs) and investigate the presence of antibiotic-resistant genes in the COVID-19 era in Northern Iran.
Methods
This cross-sectional research across four teaching hospitals Bou Ali Sina, Fatemeh Zahra, Imam Khomeini and Shahid Zare in Sari, Iran, between May 2022 and June 2023. The research aimed to evaluate the incidence of MDR and XDR P. aeruginosa among patients with NIs, based on the national directory of NIs surveillance system. The study included various samples of hospitalized patients with NIs who contracted a new infection after three days of hospitalization. Additionally, it specifically focused on including MDR P. aeruginosa among the gram-negative isolates. Outpatients, as well as gram-positive and gram-negative isolates without multiple resistance, were excluded.
A laboratory technician collected various samples from 340 patients who were suffering from HAIs. The samples included urine, sputum, wound swabs, blood, cerebrospinal fluid (CSF), pleural fluid, eye swabs, endotracheal tube (ETT) samples, ear swabs and ascites. The samples were immediately sent to the central microbiology laboratory for analysis. As part of the routine procedure, all samples were cultured on MacConkey and blood agar (QUELAB, USA) plates. Blood specimens were inoculated in BD BACTEC plus Aerobic/F culture bottles (Becton Dickinson Company, Ireland). These bottles were then incubated in a BACTEC FX40 (BD, USA) culture system. Standard microbiological procedures were followed to identify P. aeruginosa [4, 5].
Demographic data were collected from the patients’ files (Table 1).

To test P. aeruginosa susceptibility, the standard broth dilution technique was used. Macro dilution trays were prepared with reagent-grade powders from manufacturers. The MIC of ampicillin-sulbactam, ceftazidime, cefepime, ciprofloxacin, colistin, co-amoxiclav, gentamicin, meropenem and piperacillin-tazobactam (Sigma, Germany) was determined to evaluate their efficacy. Strains resistant to at least one agent in three or more antimicrobial classes were described as MDR, while strains resistant to at least one agent in all but two or fewer antimicrobial classes were denoted as XDR. To combat the spread of antibiotic resistance, strains that produced ESBLs using the double-disk synergy test were screened. At least one of the following cephalosporins was used: Cefotaxime, cefepime, ceftazidime and ceftriaxone (Padtan Teb, Iran) to identify ESBL-producing strains. Additionally, the presence of ESBLs was detected using ceftazidime/clavulanic acid (30/10 μg), cefotaxime (30 μg), cefotaxime/clavulanic acid (30/10 μg), and cefepime/clavulanic acid (30/10 μg) antibiotic disks (Padtan Teb, Iran). The P. aeruginosa ATCC 27853 strain served as the positive control [6].
According to the company’s instructions, the DNA of bacteria was extracted using a commercial DNA extraction kit (Yekta Tajhiz, Iran). All isolates were screened by the multiplex PCR method for AmpC beta-lactamases, ESBL genes (blaCTX and blaSHV), MBL genes (blaSIM, blaSPM, and blaIMP), as well as ExoA and ExoS using specific primers as previously defined 4-5. The list of primers is presented in Table 1. The multiplex PCR reaction was prepared in a final volume of 20 μL, which included 10 μL of taq DNA polymerase 2x master mix RED, 1.5 mM of MgCL2 (AMPLIQON, Denmark), 0.5 μL of each primer (10 PM), 2 μL of DNA template (100 ng) and DNase-free distilled water. The amplification was performed using a Touchdown (TD) PCR protocol [7]. The following strains were used as positive controls: Klebsiella pneumoniae ATCC NO.51503 (blaCTX-M), K. pneumoniae ATCC NO. 700603 (blaSHV), Escherichia coli ATCC BAA NO.1143 (blaAmpC), Escherichia coli NCTC NO. 13476 (blaIMP), P. aeruginosa NCTC NO. 13921 (blaSPM) and P. aeruginosa NCTC NO. 14361 (blaSIM).
Statistical analysis
SPSS software, version 22 was used to analyze the data, employing descriptive statistics, chi-square tests, and Fisher’s exact tests.
Results
Out of 340 hospitalized patients with NIs caused by drug-resistant gram-negative bacterial pathogens, MDR and XDR P. aeruginosa were responsible for 104 cases (30.6%) of NIs. The median age of the patients was 58 years (IQR: 27–69.25 years), of whom 68 (65.4%) were male and 36 (34.6%) were female. The most common NIs caused by P. aeruginosa were pneumonia (39 cases; 37.5%), UTI (36 cases; 34.6%), surgical site infection (SSI) (22 cases; 21.2%) and bloodstream infection (BSI) (7 cases; 6.7%) (Table 1). The incidence of different types of NIs caused by MDR P. aeruginosa in various hospitalization wards was significantly different, as follows: ICU (52 cases; 50%), surgery (15 cases; 14.4%), general (14 cases; 13.5%), pediatric intensive care unit (PICU) (13 cases; 12.5%), emergency (7 cases; 6.7%) and pediatrics (3 cases; 2.9%) (P=0.002). The NIs caused by MDR and XDR P. aeruginosa in different wards are shown in Figure 1. Notably, 21.2% of strains isolated from patients with NIs (40.9% pneumonia, 40.9% UTIs, 13.6% SSIs and 4.5% BSIs) exhibited an XDR phenotype. Overall, 90.4% of isolates were ESBL-producing P. aeruginosa. Table 2 summarizes the in vitro antibiotic susceptibilities, including the MIC50, MIC90, geometric means (GM) MIC, and mode of MICs for ampicillin-sulbactam, ceftazidime, cefepime, ciprofloxacin, colistin, co-amoxiclav, gentamicin, meropenem and piperacillin-tazobactam against MDR and XDR P. aeruginosa.

In terms of MIC50 values, co-amoxiclav, and gentamicin exhibited the lowest activity against P. aeruginosa. Regarding the geometric mean of minimum inhibitory concentration (GM MIC)values, meropenem exhibited the most potent activity, while co-amoxiclav displayed the lowest activity against most P. aeruginosa isolates. Also, the incidence of ESBL-producing P. aeruginosa among MDR and XDR isolates was 88.1% and 100%, respectively.
MIC50 and MIC90 represent the concentrations required to inhibit 50% and 90% of the isolates, respectively, while GM refers to the geometric mean.
All isolates encoded ExoS gene, while the gene encoding ExoA was not observed among the isolates. The frequencies of resistance genes were as follows: blaSHV (91. 3%), blaCTX-M (76%), blaAmpC(91.3%) and blaIMP (95.2%), respectively. The blaSPM and blaSIM were not detected among the isolates. Based on antibiotic-resistant genes, seven genotypes were observed among the isolates. All isolates contained at least two antibiotic-resistant encoding genes. The most common genotypes included the co-presence of blaSHV, blaCTX-M, blaAmpC and blaIMP in 60 isolates (57.7%), blaSHV, blaAmpCand blaIMP in 25 isolates (24%), blaCTX-M, blaAmpC and blaIMP in eight isolates (7.7%), blaSHV, blaCTX-M and blaIMP in five isolates (4.8%), blaSHV, blaCTX-M, blaAmpCin three isolates (2.9%), blaSHV and blaAmpC in two isolates (1.9%), blaCTX-M and blaIMP in one isolate (1%), respectively. The blaCTX-M gene was significantly associated with resistance to ciprofloxacin (P<0.006) and the blaIMP gene was significantly associated with resistance to meropenem (P<0.03) (Tables 3 and 4).


 #T4
#T4Discussion
P. aeruginosa is a major challenge in hospitals, causing high mortality rates, prolonged stays and increased costs [1, 2, 3]. The current study was carried out in the northern region of Iran and found that P. aeruginosa was accountable for 30.6% of NIs caused by MDR pathogens, with pneumonia accounting for the majority of these infections at 37.5%. Interestingly, previous surveillance studies conducted in teaching hospitals in the same area prior to the COVID-19 pandemic showed a significantly lower incidence of nosocomial pneumonia caused by ESBL-producing P. aeruginosa, at 14.63% [8]. Before the COVID-19 pandemic, UTIs (26.8%), VAP (20.3%), SSIs (19.7%) and BSIs (13.5%) were the most common NIs in different regions of Iran, according to a 2020 study [9]. The high occurrence of pneumonia in the current research is attributed to the high rate of ICU and PICU patients requiring invasive and non-invasive ventilation.
According to a 2014 survey, Pseudomonas spp. was the most commonly found bacteria in wound infections (50.81%), respiratory infections (21.31%), UTIs (19.67%) and blood infections (8.19%) [10]. However, a recent survey conducted in the post-COVID-19 era revealed that UTIs (33.33%), respiratory infections (32.46%), wound infections (19.30%) and blood infections (6.14%) were the most prevalent types of infections. In our study, MDR and XDR P. aeruginosa were responsible for 34.6% of UTIs, which is alarming because these infections are linked to severe forms of UTIs. They are often associated with urinary catheter-related infections, prostatitis and urolithiasis, which are very challenging to treat. Although P. aeruginosa is the third most common pathogen linked to nosocomial UTIs, the high incidence of UTIs caused by this bacterium is of particular concern [11].
SSIs account for 15% of all NIs in surgical patients, and SSIs caused by MDR bacteria are associated with significant postoperative complications and preoperative surgery [12]. The most common causes of SSI are Staphylococcus spp., Enterococcus spp., Streptococcus spp., and Pseudomonas spp. [13]. In this research, SSIs were caused by MDR and XDR P. aeruginosa in 22.2% of cases, consistent with another research [14]. Understanding the development and risk factors of SSI can help reduce treatment costs and allocate resources efficiently. In our study, the rate of BSI caused by MDR and XDR P. aeruginosa was 6.7%. There was also a link between AMR and poor clinical outcomes in patients with BSIs. Specifically, patients infected with MDR P. aeruginosa have twice the odds of mortality in comparison to those with non-MDR P. aeruginosa infections [15].
In a previous study involving patients with VAP and sepsis, it was discovered that approximately half of the isolated P. aeruginosa bacteria were resistant to all aminoglycoside antibiotics and 45.85% were resistant to ciprofloxacin. Moreover, the study found that P. aeruginosa exhibited resistance rates of 62.5% to colistin and 29.2% to imipenem [3]. After the COVID-19 outbreak, our recent surveillance investigations revealed an enhancement in resistance to aminoglycoside antibiotics among isolated P. aeruginosa bacteria, reaching 76%. Additionally, resistance to ciprofloxacin has risen to 77.9%. The resistance rates to colistin and meropenem among P. aeruginosa were found to be 70.2% and 50%, respectively.
Our results (2017) revealed a high incidence of catheter-associated UTIs in hospitals [16]. P. aeruginosa was recognized as a major cause of UTIs and demonstrated resistance to aminoglycosides (56%), fluoroquinolones (63%) and third-generation cephalosporins (38%). Following the COVID-19 outbreak, a further increase was observed in resistance to aminoglycosides (68.42%), fluoroquinolones (71.05%) and third-generation cephalosporins (68.42%) based on UTI samples. Tiri et al. found a significant increase in carbapenem-resistant Enterobacterales colonization from 6.7% (2019) to 50% (2020) [17].
During the pandemic period, AlDiba et al. discovered a substantial increase in the prevalence of carbapenem-resistant Enterobacterales, which rose to 22.4% from 5.4% in the pre-pandemic period [18]. The CDC’s latest report shows a 35% increase in MDR P. aeruginosa infections from 2019 to 2020, possibly due to the high administration of antibiotics to treat secondary bacterial infections associated with SARS-CoV-2 [19].
The rational usage of antibiotics during the COVID-19 pandemic was not followed. The antibiotics prescribed for COVID-19 patients were higher than the incidence of bacterial co-infections in these patients (bacterial co-infection [3.5%] and secondary bacterial infection [14.3%]) [20]. The high use of antibiotics in patients with COVID-19 during the pandemic raises concerns about the emergence of antimicrobial-resistant pathogens, such as resistant P. aeruginosa species, which have developed resistance to several antibiotic categories, such as cephalosporins, carbapenems, or polymyxins [1, 20]. In this study, a high resistance phenotype (38.5-100%) to different classes of antibiotics was observed, which is significantly higher than our previous finding (26-86.66%) before COVID-19 [3]. In the current study, piperacillin-tazobactam and imipenem were the most effective antibiotics against MDR and XDR P. aeruginosa. Our findings were in agreement with those of Ahmadi et al. who also reported that MDR P. aeruginosa was most susceptible to meropenem and piperacillin [21]. The antimicrobial sensitivity pattern in our examined strains showed higher resistance than P. aeruginosa isolates in other studies. This difference may be related to the studied population, the type of infections (as all strains were isolated from patients with NIs, not community-acquired infections) and the study period of three years after the COVID-19 pandemic.
The frequency of different ESBL genotypes varies in different regions. The fact that 57.6% of isolates in the current investigation contain all antibiotic-resistant genes (AmpC beta-lactamases, ESBL genes and MBL genes) is worrying. Several studies have reported the simultaneous presence of different β-lactamase genes in the same strains [22, 23]. The acquisition of drug-modifying enzymes in P. aeruginosa, such as ESBL and carbapenemases, can be accessed through horizontal gene transfer [4, 5]. Consistent with the current study, the blaSHV gene (86.66%) was the most detected ESBL gene in our previous survey [3]. Similar studies on ESBL-encoding genes revealed diverse findings [24]. The blaTEM, blaCTX and blaSHV variants have been the most common ESBLs during the past decade [25-27]. In this study, the most antibiotic-resistant-encoding gene was blaIMP, detected in 95.2% of isolates. However, 39.4% of isolates contained the blaIMP gene and showed a susceptible phenotype to meropenem. The incidence of blaIMP gene and resistance to meropenem was statistically significant (P=0.037). Most ESBL-encoding genes are plasmid-mediated enzymes, which are transmitted easily among bacteria, leading to inappropriate or failed antimicrobial therapy [28, 29]. Constant monitoring of bacteria carrying antibiotic resistance encoding genes (ESBL and MBL-producing strains) is pivotal to designating appropriate antimicrobial therapy.
Certain limitations in our study need to be taken into account. Firstly, the rate of hospital-acquired infections caused by MDR and XDR P. aeruginosa, as well as the presence of resistance genes, might have been underestimated due to partial treatment before the bacterial cultures were obtained in some cases. Secondly, we did not sequence the following genes associated with resistance phenotypes: blaSHV, blaCTX-M, blaAmpC and blaIMP, which limited our ability to match in vitro and in vivo resistance results. Despite these limitations, our findings provide crucial insights.
Conclusion
Our findings showed that UTIs, sputum and blood samples have the highest prevalence of blaSHV, blaCTX-M, blaAmpC and blaIMP genes in MDR P. aeruginosa isolates. The presence of these genes underscores the urgent need to expand empiric antibiotic therapy for critically ill patients. In addressing hospital-acquired infections caused by MDR bacteria, like carbapenemase and ESBL producers, it is imperative to implement suitable empiric or alternative treatments based on epidemiological data or relevant antibiotic exposure history. This study recommends the experimental use of meropenem to treat NIs caused by MDR P. aeroginosa in hospitals located in northern Iran.
Ethical Considerations
Compliance with ethical guidelines
This study was approved by the Ethics Committee of Mazandaran University of Medical Sciences (Code: IR. MAZUMS.REC.1401.13944).
Funding
This research did not receive any grant from funding agencies in the public, commercial, or non-profit sectors.
Authors contributions
All authors equally contributed to preparing this article.
Conflicts of interest
The authors declared no conflicts of interest.
Acknowledgements
The authors gratefully acknowledge Fatemeh Bagherzadeh, Ebrahim Nemati and Shahram Divsalar for their supportive assistance during the laboratory tests.
References
- Sreenath K, Batra P, Vinayaraj E, Bhatia R, SaiKiran K, Singh V, et al. Coinfections with other respiratory pathogens among patients with COVID-19. Microbiol Spectr. 2021; 9(1):e00163-21. [DOI:10.1128/Spectrum.00163-21]
- Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019; 32(4):10-128. [DOI:10.1128/CMR.00031-19]
- Rezai MS, Ahangarkani F, Rafiei A, Hajalibeig A, Bagheri-Nesami M. Extended-spectrum beta-lactamases producing Pseudomonas aeruginosa isolated from patients with ventilator associated nosocomial infection. Arch Clin Infect Dis. 2018; 13(4):e13974. [DOI:10.5812/archcid.13974]
- Rahimzadeh G, Rezai MS. [Detection extended-spectrum beta-lactamase-and carbapenemase-producing enterobacteriaceae isolates from clinical samples; Narrative review (Persian)]. J Isfahan Med Sch. 2022; 40:743-58. [DOI:10.48305/jims.v40.i688.0743]
- Rahimzadeh G, Rezai M, Farshidi F. Genotypic patterns of multidrug resistant Acinetobacter baumannii: A systematic review. Adv Biomed Res. 2022; 12(1):56. [DOI:10.4103/abr.abr_434_22]
- Rahimzadeh G, Valadan R, Rezai S, Khosravi M, Larijani LV, Sheidaei S, et al. Evaluation of antibiotic resistance changes in Acinetobacter baumannii in the era of COVID-19 in Northern Iran. Iran J Microbiol. 2024; 16(3):314-22. [DOI:10.18502/ijm.v16i3.15762]
- Korbie DJ, Mattick JS. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat Protoc. 2008; 3:1452-6. [DOI:10.1038/nprot.2008.133]
- Bagheri-Nesami M, Rezai MS, Ahangarkani F, Rafiei A, Nikkhah A, Eslami G, et al. Multidrug and co-resistance patterns of non-fermenting Gram-negative bacilli involved in ventilator-associated pneumonia carrying class 1 integron in the North of Iran. Germs. 2017; 7(3):123-31. [DOI:10.18683/germs.2017.1117] [PMID]
- Tafti A, Zandi H, Vakli M, Mousavi SM, Zarei M. [Frequency of β-lactamase and metallo-β-lactamase in Pseudomonas aeruginosa strains isolated from burn wounds in Yazd burn hospital during 2011-2012 (Persian)]. Feyz. 2014; 18(2):167-74. [Link]
- Behzadnia S, Davoudi A, Rezai MS, Ahangarkani F. Nosocomial infections in pediatric population and antibiotic resistance of the causative organisms in north of iran. Iran Red Crescent Med J. 2014; 16(2):e14562. [DOI:10.5812/ircmj.14562] [PMID]
- Rahimzadeh M, Habibi M, Bouzari S, Asadi Karam MR. First study of antimicrobial activity of ceftazidime-avibactam and ceftolozane-tazobactam against Pseudomonas aeruginosa isolated from patients with urinary tract infection in Tehran, Iran. Infecti Drug Resist. 2020; 2020:533-41. [DOI:10.2147/IDR.S243301]
- Foschi D, Yakushkina AO, Cammarata F, Lamperti G, Colombo F, Rimoldi S, et al. Surgical site infections caused by multi-drug resistant organisms: A case–control study in general surgery. Updat Surg. 2022; 74(5):1763-71. [DOI:10.1007/s13304-022-01243-3]
- Puca V, Marulli RZ, Grande R, Vitale I, Niro A, Molinaro G, Pet al. Microbial species isolated from infected wounds and antimicrobial resistance analysis: Data emerging from a three-years retrospective study. Antibiotics. 2021; 10(10):1162. [DOI:10.3390/antibiotics10101162]
- Heidari R, Farajzadeh Sheikh A, Hashemzadeh M, Farshadzadeh Z, Salmanzadeh S, Saki M. Antibiotic resistance, biofilm production ability and genetic diversity of carbapenem-resistant Pseudomonas aeruginosa strains isolated from nosocomial infections in southwestern Iran. Mol Biol Rep. 2022; 49(5):3811-22. [DOI:10.1007/s11033-022-07225-3]
- Recio R, Mancheño M, Viedma E, Villa J, Orellana MÁ, Lora-Tamayo J, et al. Predictors of mortality in bloodstream infections caused by Pseudomonas aeruginosa and impact of antimicrobial resistance and bacterial virulence. Antimicrob Agents Chemother. 2020; 64(2):10-128. [DOI:10.1128/AAC.01759-19]
- Rezai MS, Bagheri-Nesami M, Nikkhah A. Catheter-related urinary nosocomial infections in intensive care units: An epidemiologic study in North of Iran. Caspian J Intern Med. 2017; 8(2):76. [DOI:10.22088/cjim.8.2.76] [PMID]
- Tiri B, Sensi E, Marsiliani V, Cantarini M, Priante G, Vernelli C, et al. Antimicrobial stewardship program, COVID-19, and infection control: Spread of carbapenem-resistant Klebsiella pneumoniae colonization in ICU COVID-19 patients. What did not work?. J Clin Med. 2020; 9(9):2744. [DOI:10.3390/jcm9092744]
- AlDiba M, Daghriri AM, Jamali EM, Alzahrani A, Alsharif AB, Almudeer HG, et al. Prevalence of antimicrobial resistance of common bacterial isolates before and during COVID-19 pandemic in armed forces hospital Jazan, Saudi Arabia. Eur J Med Health Sci. 2021; 3(5):31-8. [DOI:10.24018/ejmed.2021.3.5.1047]
- Langford BJ, So M, Simeonova M, Leung V, Lo J, Kan T, et al. Antimicrobial resistance in patients with COVID-19: A systematic review and meta-analysis. Lancet Microbe. 2023; 4(3):e179-91. [DOI:10.1016/S2666-5247(22)00355-X]
- Langford BJ, So M, Raybardhan S, Leung V, Soucy JP, Westwood D, et al. Antibiotic prescribing in patients with COVID-19: Rapid review and meta-analysis. Clin Microbiol Infect. 2021; 27(4):520-31. [DOI:10.1016/j.cmi.2020.12.018]
- Ahmadi N, Salimizand H, Zomorodi AR, Abbas JE, Ramazanzadeh R, Haghi F, et al. Genomic diversity of β-lactamase producing Pseudomonas aeruginosa in Iran; the impact of global high-risk clones. Ann Clin Microbiol Antimicrob. 2024; 23(1):5. [DOI:10.1186/s12941-024-00668-5]
- Hosu MC, Vasaikar SD, Okuthe GE, Apalata T. Detection of extended spectrum beta-lactamase genes in Pseudomonas aeruginosa isolated from patients in rural Eastern Cape Province, South Africa. Sci Rep. 2021; 11(1):7110. [DOI:10.1038/s41598-021-86570-y]
- Zafer MM, Al-Agamy MH, El-Mahallawy HA, Amin MA, Ashour MS. Antimicrobial resistance pattern and their beta-lactamase encoding genes among pseudomonas aeruginosa strains isolated from cancer patients. Biomed Res Int. 2014; 2014(1):101635. [DOI:10.1155/2014/101635]
- Finkelstein R, Rabino G, Mashiah T, Bar-El Y, Adler Z, Kertzman V, et al. Surgical site infection rates following cardiac surgery: The impact of a 6-year infection control program. Am J Infect Control. 2005; 33(8):450-4. [DOI:10.1016/j.ajic.2005.07.002]
- Chen Z, Niu H, Chen G, Li M, Li M, Zhou Y. Prevalence of ESBLs-producing Pseudomonas aeruginosa isolates from different wards in a Chinese teaching hospital. Int J Clin Exp Med. 2015; 8(10):19400-5. [PMID]
- Maurya AP, Talukdar AD, Chanda DD, Chakravarty A, Bhattacharjee A. Integron-borne transmission of VEB-1 extended-spectrum β-lactamase in Pseudomonas aeruginosa in a tertiary care hospital in India. Antimicrob Agents Chemother. 2014; 58(11):6966-9. [DOI:10.1128/AAC.02365-14]
- Lewis JS, Herrera M, Wickes B, Patterson JE, Jorgensen JH. First report of the emergence of CTX-M-type extended-spectrum β-lactamases (ESBLs) as the predominant ESBL isolated in a US health care system. Antimicrob Agents Chemother. 2007; 51(11):4015-21. [DOI:10.1128/AAC.00576-07]
- Rajaee Behbahani M, Keshavarzi A, Pirbonyeh N, Javanmardi F, Khoob F, Emami A. Plasmid-related β-lactamase genes in Pseudomonas aeruginosa isolates: A molecular study in burn patients. J Med Microbiol. 2019; 68(12):1740-6. [DOI:10.1099/jmm.0.001105]
- Kurittu P, Khakipoor B, Aarnio M, Nykäsenoja S, Brouwer M, Myllyniemi AL, Vatunen E, et al. Plasmid-borne and chromosomal ESBL/AmpC genes in Escherichia coli and Klebsiella pneumoniae in global food products. Front Microbiol. 2021; 12:592291. [DOI:10.3389/fmicb.2021.592291]
Type of Study: Original Article |
Subject:
Microbiology
Received: 2024/06/20 | Accepted: 2024/09/1 | Published: 2024/07/31
Received: 2024/06/20 | Accepted: 2024/09/1 | Published: 2024/07/31
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |